How to Read Multiple Excel or CSV Files Together
You might have multiple Excel or CSV files that share the same data structure (same columns) and are stored in the same folder. If these are only a few you can import them one by one and bind them together with ‘bind_rows’ command in Exploratory.
But if there are tons, that’s not really a reasonable option. What should we do?
Read Multiple Files in R
In R, you can write a script to read all the files in the same folder and bring them relatively easily.
Let’s say we have three Excel files under this folder.
Here is a script for reading them together.
library(readxl)
files <- list.files(path = "~/Dropbox/Data/multiple_files", pattern = "*.xlsx", full.names = T)
tbl <- sapply(files, read_excel, simplify=FALSE) %>%
bind_rows(.id = "id")If you have multiple CSV files instead of Excel files, here is one for you.
library(readr)
files <- list.files(path = "~/Dropbox/Data/multiple_files", pattern = "*.csv", full.names = T)
tbl <- sapply(files, read_csv, simplify=FALSE) %>%
bind_rows(.id = "id") Only the difference is either using ‘read_csv’ function from ‘readr’ package for reading CSV files or using ‘read_excel’ function from ‘readxl’ package for reading Excel files.
Either way, this is what’s happening with the above scripts.
- Tries to find all the files whose names ending with ‘xlsx’ or ‘csv’ and store the file location information into ‘files’ variable.
- Store the file names into ‘files_df’ data frame.
- Read the files one by one and bind them together.
- Finally, it joins the data frame that combined the multiple files with the data frame with file names (files_df).
Use Custom R Script as Data Source in Exploratory
If you can write an R script that means you can make the script as a data source in Exploratory.
Select ‘R Script’ from the data frame menu.
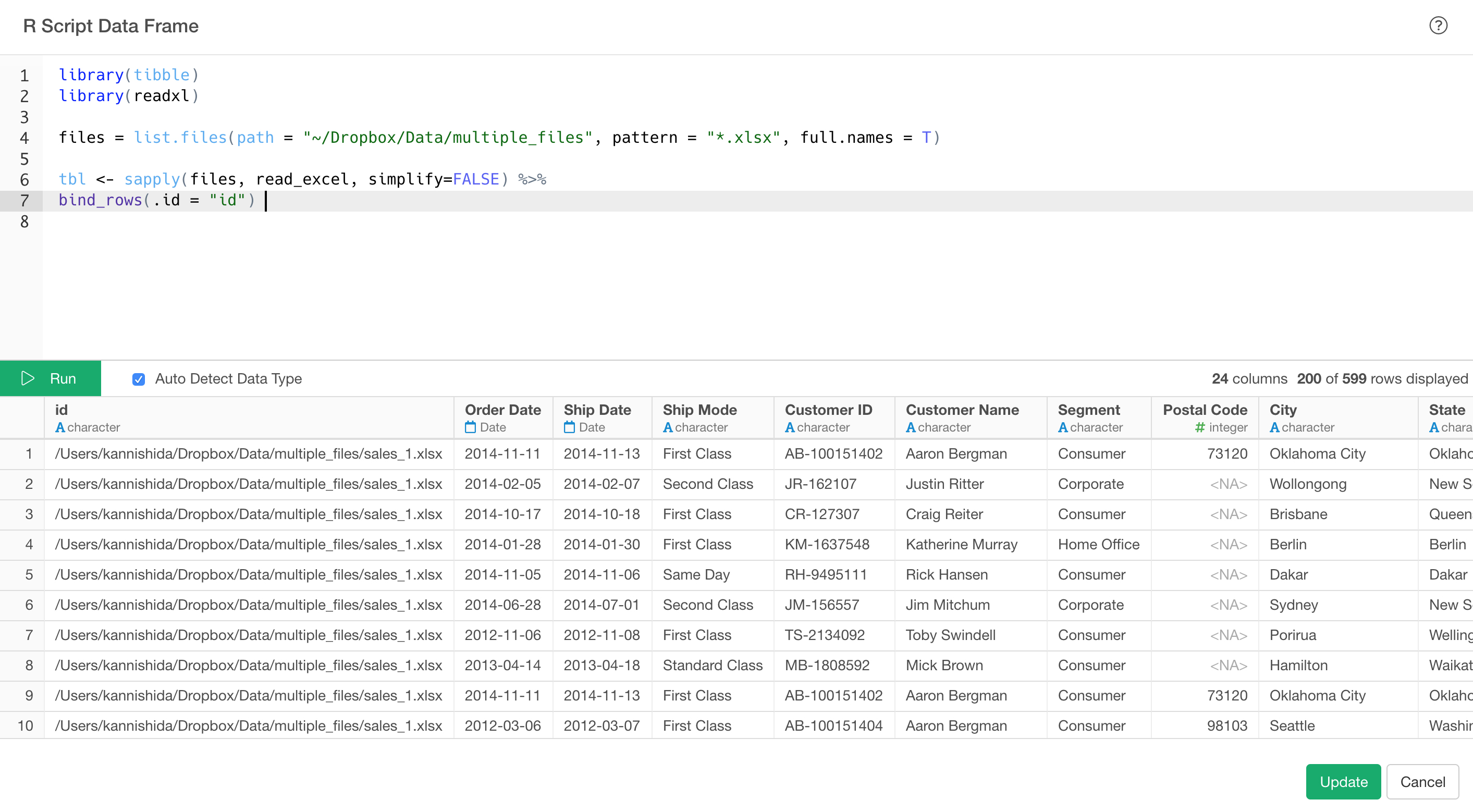
In the R Script Editor, you can copy and paste the above script. Here, I’m pasting the script for reading multiple Excel files.
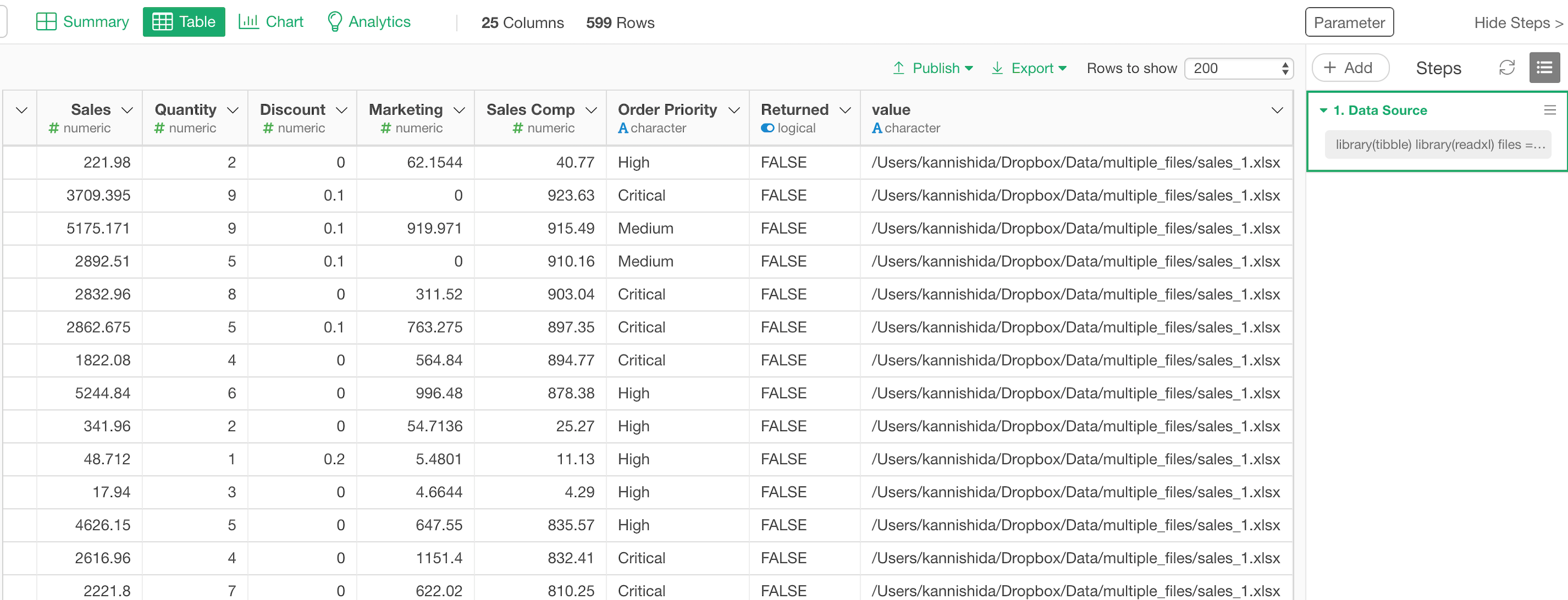
Once you click on ‘Save’ button the multiple files are imported together and you will have one single data frame.
You can see the original file paths in the last column.
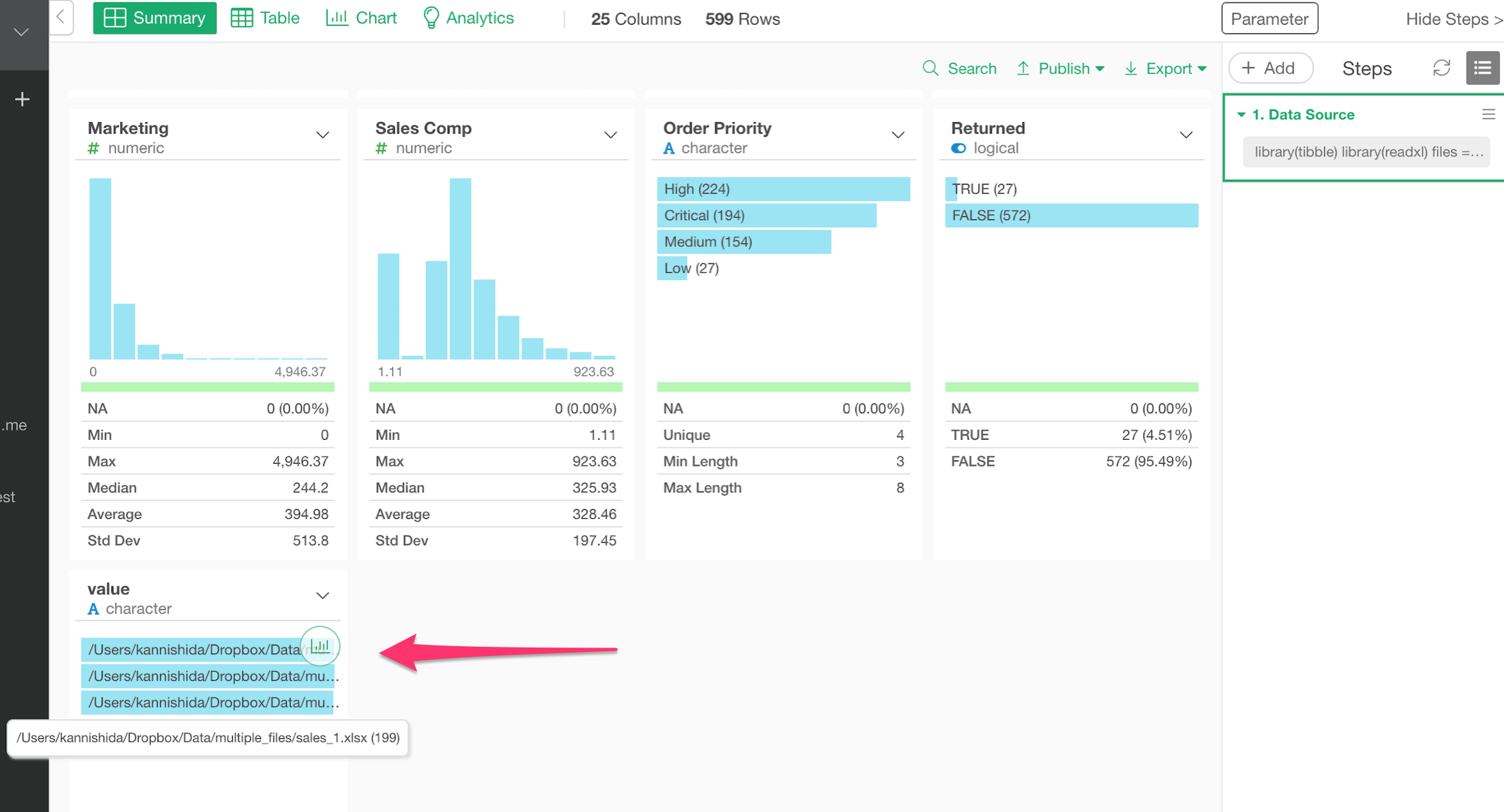
Here’s how it looks in Table view.